New preprint from our lab: Global distribution of quinones in Bacteria!
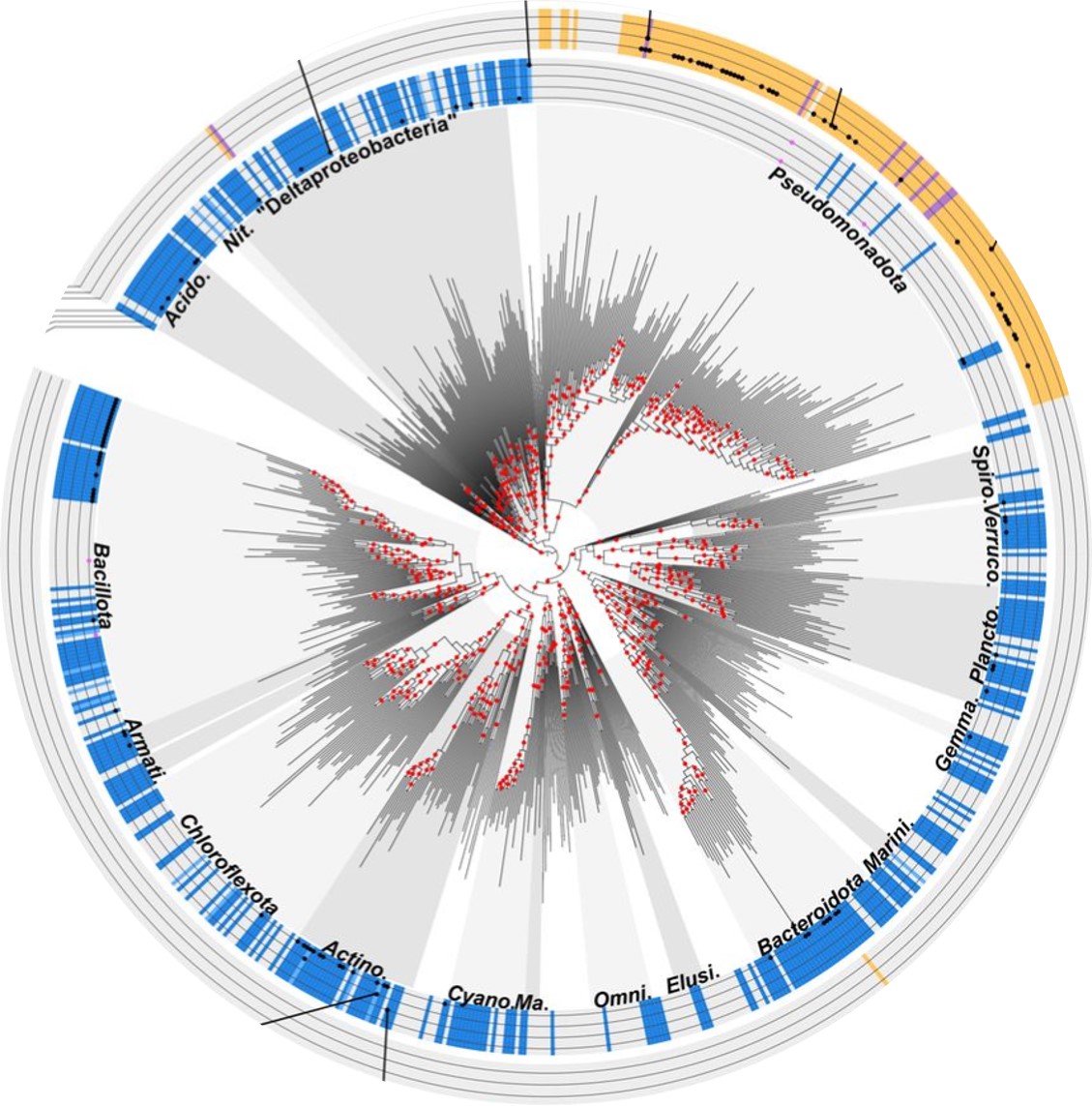
Really glad to announce a new preprint from our lab (TrEE team): Global distribution of quinones in Bacteria!
👉 https://www.biorxiv.org/content/10.1101/2025.09.17.676790v1
We here combine comparative genomics and text mining to leverage information on quinones distribution from thousands of articles describing new species and from dozens of thousands of genomes!
Among other discoveries, we show that the two menaquinone pathways tend to replace one another frequently, and that a lineage outside of Pseudomonadota unexpectedly show the potential to produce ubiquinone, a high redox potential usually found in Pseudomonadota and Eukaryota.
This much-needed compendium of quinones in bacteria is the last chapter of S. Chobert's PhD thesis. This comes after the discovery of a high potential quinone, the methylplastoquinone, led by Felix Elling (Micro-Bio-Geologist) and colleagues published earlier in PNAS (Proceedings of the National Academy of Sciences) and after the study brilliantly conducted by Dr. Chobert on the dynamics of the quinone repertoire in Pseudomonadota that accompanied their metabolic diversification, also published earlier this year in the ISME Journal from ISME (International Society for Microbial Ecology).
We would like to thank the scientists across the world who characterize quinones in newly described species and publish their work e.g. in IJSEM from Microbiology Society, Antonie van Leeuwenhoek, Archives of Microbiology and Systematic and Applied Microbiology.
Quinones rock!
